In a previous post, we discussed how the logic of genetic mapping is based on counting the new genetic combinations that appear in the offspring of a cross as a result of crossover events during meiosis.
Today, let’s look at the standard bi-parental populations (mapping populations) used for this purpose.
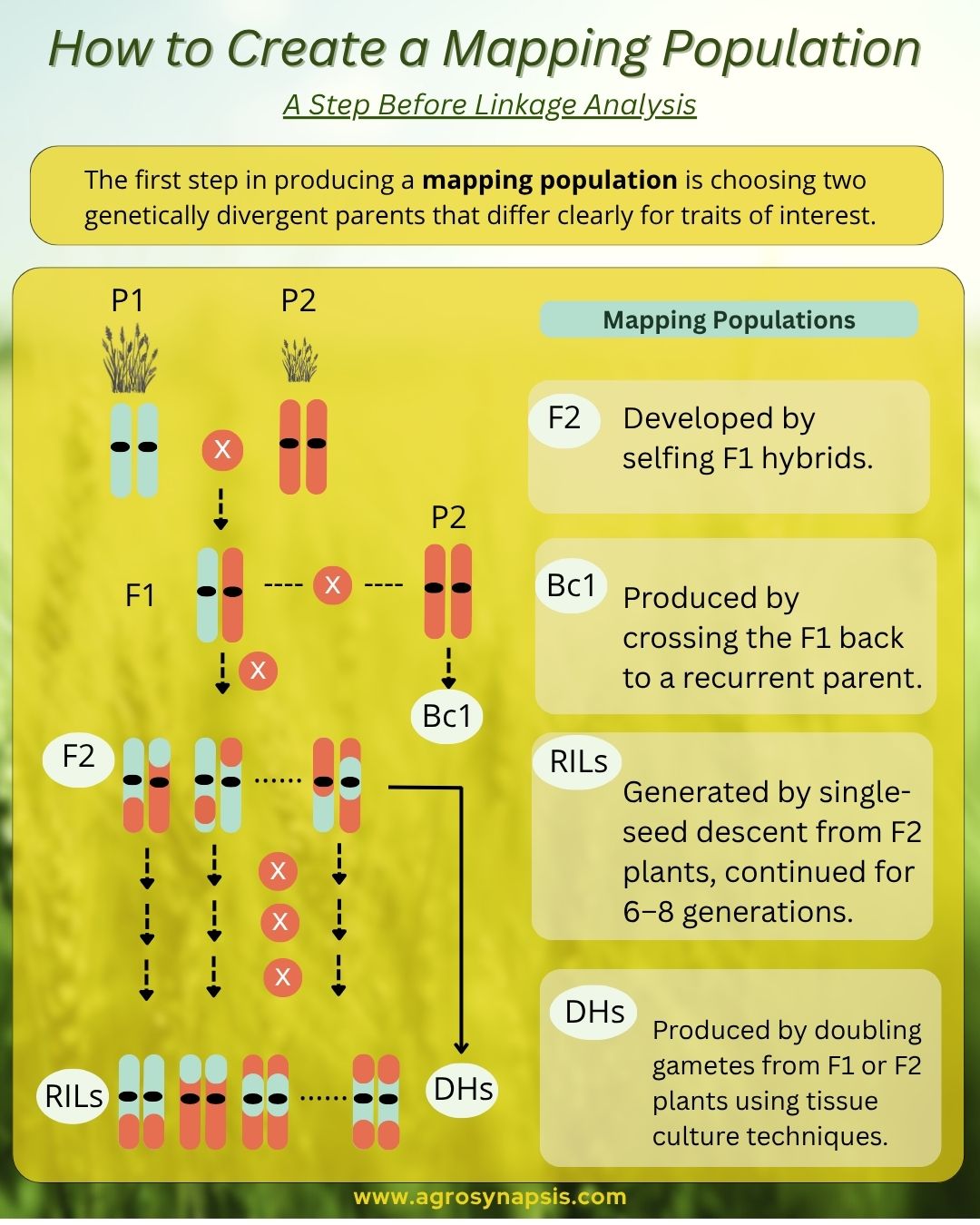
The first step in producing a mapping population is choosing two genetically divergent parents that differ clearly for traits of interest. The right balance matters: they should show enough genetic variation to reveal polymorphisms, but not be so distant that progenies suffer from sterility or extreme segregation distortion.
👉 In self-pollinating species, mapping populations are usually derived from inbred parents.
👉 In cross-pollinating species, things are more complex—since most don’t tolerate inbreeding, population design requires special care.
Types of Mapping Populations
1️⃣ F2 populations: developed by selfing F1 hybrids.
2️⃣ Backcross (BC) populations: produced by crossing the F1 back to a recurrent parent.
3️⃣ Recombinant Inbred Lines (RILs): generated by single-seed descent from F2 plants, continued for 6–8 generations.
4️⃣ Double Haploids (DHs): produced by doubling gametes from F1 or F2 plants using tissue culture techniques.
🔬 Permanent populations like RILs and DHs are highly valuable because they are homozygous and can be reproduced without genetic change—making them easy to share across labs for collaborative studies and for trials with replications.
⏱️ Time vs. feasibility:
hashtag#DHs are quicker to produce than RILs, but only for species with established haploid culture protocols.
hashtag#RILs take longer but remain a cornerstone for many crops. In addition, they accumulate more recombination events during successive generations of selfing, making mapping more efficient and accurate. Their production can be accelareted by hashtag#SpeedBreeding.
📊 Population size matters: Preliminary mapping often uses 50–250 individuals, but larger populations are needed for high-resolution, fine mapping.
✨ At AgroSynapsis, we’ve just completed our first workshop on how setting up a Marker-Assisted Selection Pipeline. Participants valued the clarity and simplicity with which we explained complex concepts—something we are committed to in all our trainings and collaborations.
👉 If you’d like to be informed about our upcoming workshops, and receive early access and discounts, f𝗶𝗹𝗹 𝗼𝘂𝘁 𝗼𝘂𝗿 𝘀𝗵𝗼𝗿𝘁 𝘁𝗿𝗮𝗶𝗻𝗶𝗻𝗴 𝗶𝗻𝘁𝗲𝗿𝗲𝘀𝘁 𝗳𝗼𝗿𝗺 here:
https://lnkd.in/g3tApqPz
How to create Mapping Populations
by

